What Will Genomics Mean for You?
3 Mar 2021|Jenny Taylor
- Research
We are all very familiar with the phrase ‘It’s in our genes’ and the concept that we inherit many of our characteristics, such as aspects of our physical appearance or our mannerisms (in genetics terms known as our phenotype) from our parents. Our genetic inheritance also informs the types of diseases we may be susceptible to.
It’s in our Genes – the Basis of Human Variation
The root of this inheritance is variation in our genes that makes each of us different from the next person. This variation in turn arises from the precise order of the ‘letters’ that makes up DNA. DNA’s alphabet simply comprises four letters; A, C, G, T. But that’s where the simplicity stops. An average length of a gene is 15,000 letters and we have around 25,000 genes.
Genes and Disease
Armed now with this standard, we can explore variation; how an individual’s genome differs from the reference genome which can guide our understanding of many aspects of human biology.
Which brings us on to human disease. Most diseases have a genetic component. Some diseases are caused by variants in just one gene. These so-called monogenic conditions often have a striking pattern of inheritance in families. Some examples include Huntington’s disease, cystic fibrosis, Duchenne muscular dystrophy, sudden cardiac death syndromes and sickle cell anaemia. There are estimated to be over 8,000 such rare diseases and even if individually rare, collectively they affect 1:20 people, or 400 million people across the globe.
Whilst it may be possible to provide a clinical diagnosis for patients with these conditions, families often want to know why their children have the condition. Often parents have been obliged to go on a diagnostic odyssey looking for the cause of their children’s disease. Now with access to genomic sequencing this can be resolved quickly in a single test. So increasingly a molecular diagnosis is sought to be able answer the question ‘which gene is not working?’
To find the typo (the technical term is mutation) in a patient’s genome which is causing a rare disease is not, however, straightforward. Until recently it relied on searching painstakingly through the sequence of genes already known to cause the condition, to find the ‘typo’. Often no errors would be found because the culprit gene might not yet have been identified. But as technologies have improved and costs of sequencing have dramatically fallen, it is now feasible to sequence individual patients’ entire genomes to find the genetic basis for their rare disease.

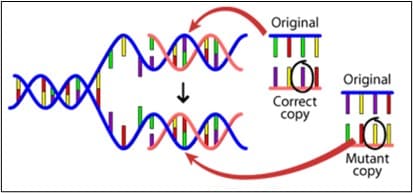
Several years ago my group conducted a large pilot study (called the WGS500 study) with the Wellcome Centre for Human Genetics to see whether genome sequencing in the clinic would be feasible. We sequenced 500 patient genomes in multiple families to establish whether it was possible to detect genetic causes of disease in many different disorders. This programme was the first to establish that genome sequencing would be of great benefit to many patients and became an important foundation for the UK’s Genomics England programme to sequence 100,000 patients.
My research group is currently involved with analysing the genome sequences for the 100,000 Genomes Project; the results have led to discovery of multiple new disease genes (two recently published examples are genes for a growth disorder and a motor neuropathy. These novel disease genes contribute to our understanding of human biology, but most importantly they give diagnoses for families affected by rare diseases.
Now, genome sequencing is commissioned by the NHS for selected rare diseases, so within 20 years of the Human Genome Project finishing, genetic testing is available on NHS for many families with rare diseases.

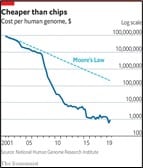
Common Diseases and Risk Prediction
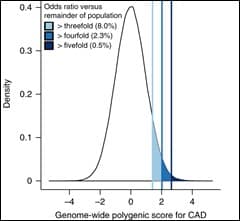
Whilst the massive investment in sequencing the human genome is now bearing fruit in rare diseases, there has been a criticism that genetics has not delivered on its promise for patients with common diseases; these are the big killers – cancer, heart disease, diabetes, stroke etc- caused by the interplay of genes and environment. We know many of the environmental factors (including smoking, cholesterol and exposure to certain chemicals) which contribute to these diseases but the definitive association of genetic factors has been less clear cut. Whilst world-wide research has identified a plethora of genetic loci associated with common diseases, the odds ratios of individual factors have not been significant enough to be used clinically.
Now, however, the cumulative impact of these risk alleles is being developed as a score to inform people of their chances of developing specific common diseases. Our group is working with many others to bring such ‘polygenic risk scores’ into general clinical practice so that we can all use the information in our own genomes to find out about our risks of the major diseases, and take preventive action accordingly.
Category: Research
Author

Jenny
Taylor
The first human genome was sequenced nearly 20 years ago, but what impact will this have for you? Understanding the information in our genes is already helping to decipher the molecular basis of rare diseases. Now genomic information is starting to be used for predicting risk of common diseases – information which could help all of us.